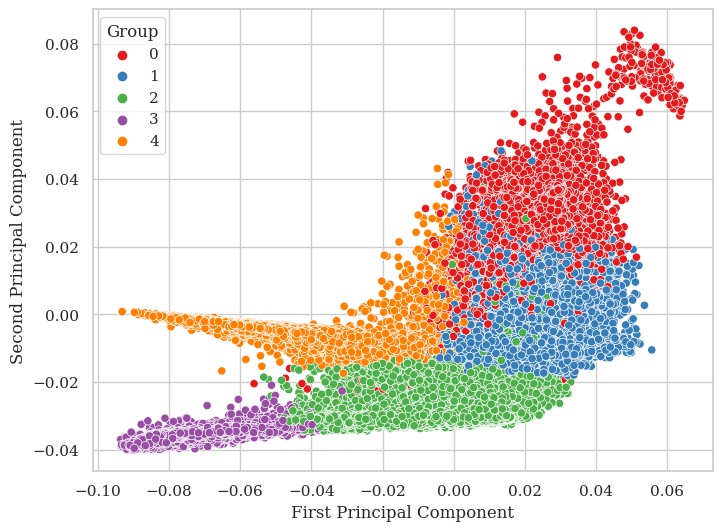
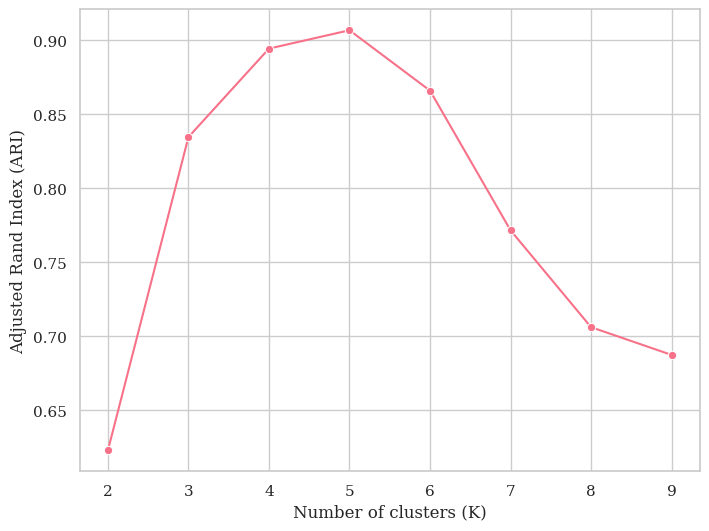
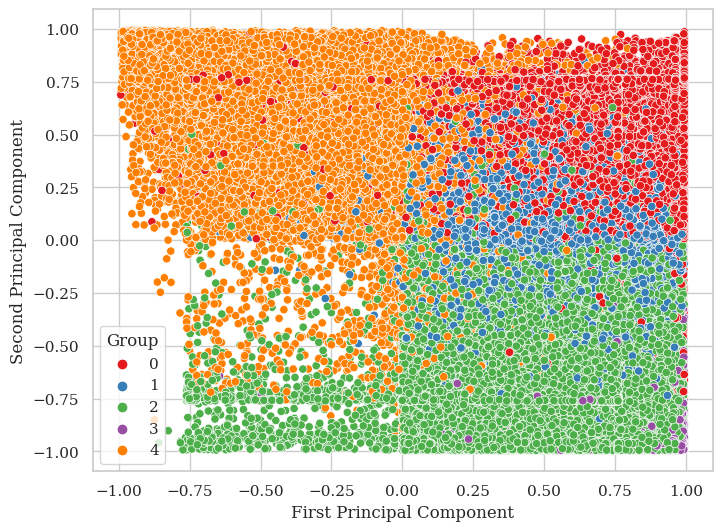
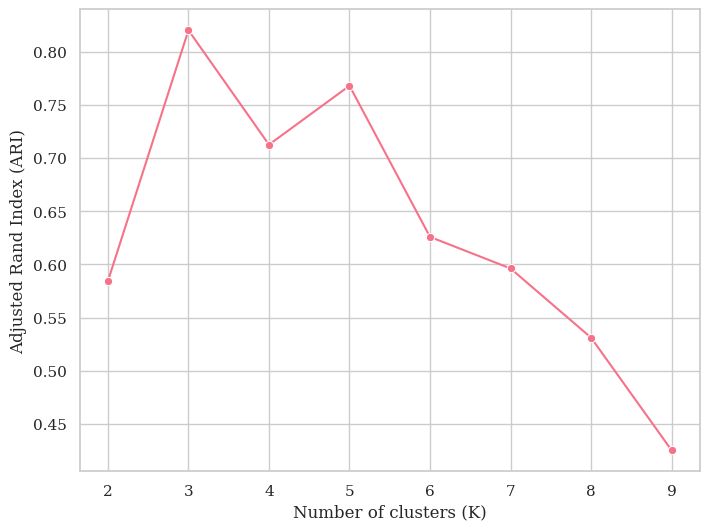
preds = np.argmax(y_train_pred, axis = 1)
true_indx = np.where(y_train_true == preds)[0]
pca = PCA(n_components=2)
principal_components = pca.fit_transform(train_projections[true_indx, :])
true_positive = y_train_true[true_indx]
df = pd.DataFrame({
'First Principal Component': train_projections[true_indx, 0],
'Second Principal Component': train_projections[true_indx, 1],
'Group': true_positive
})
plt.figure(figsize=(8, 6))
# Utilizza seaborn.scatterplot
sns.scatterplot(data=df, x='First Principal Component', y='Second Principal Component', hue='Group', palette='Set1')
plt.show()
plt.close()